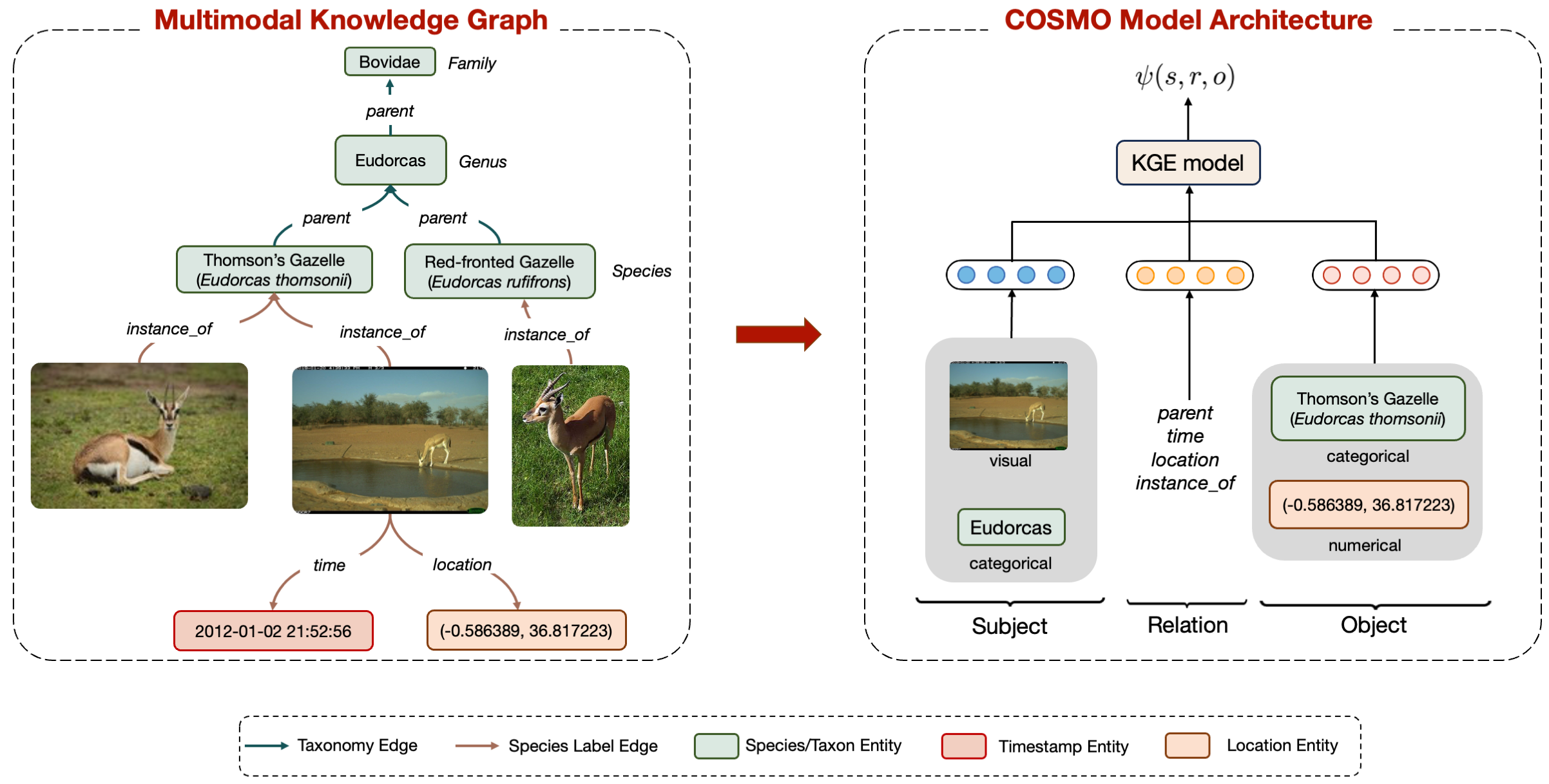
Camera traps are valuable tools in animal ecology for biodiversity monitoring and conservation. However, challenges like poor generalization to deployment at new unseen locations limit their practical application. Images are naturally associated with heterogeneous forms of context possibly in different modalities. In this work, we leverage the structured context associated with the camera trap images to improve out-of-distribution generalization for the task of species identification in camera traps. For example, a photo of a wild animal may be associated with information about where and when it was taken, as well as structured biology knowledge about the animal species. While typically overlooked by existing work, bringing back such context offers several potential benefits for better image understanding, such as addressing data scarcity and enhancing generalization. However, effectively integrating such heterogeneous context into the visual domain is a challenging problem. To address this, we propose a novel framework that reformulates species classification as link prediction in a multimodal knowledge graph (KG). This framework seamlessly integrates various forms of multimodal context for visual recognition. We apply this framework for out-of-distribution species classification on the iWildCam2020-WILDS and Snapshot Mountain Zebra datasets and achieve competitive performance with state-of-the-art approaches. Furthermore, our framework successfully incorporates biological taxonomy for improved generalization and enhances sample efficiency for recognizing under-represented species.

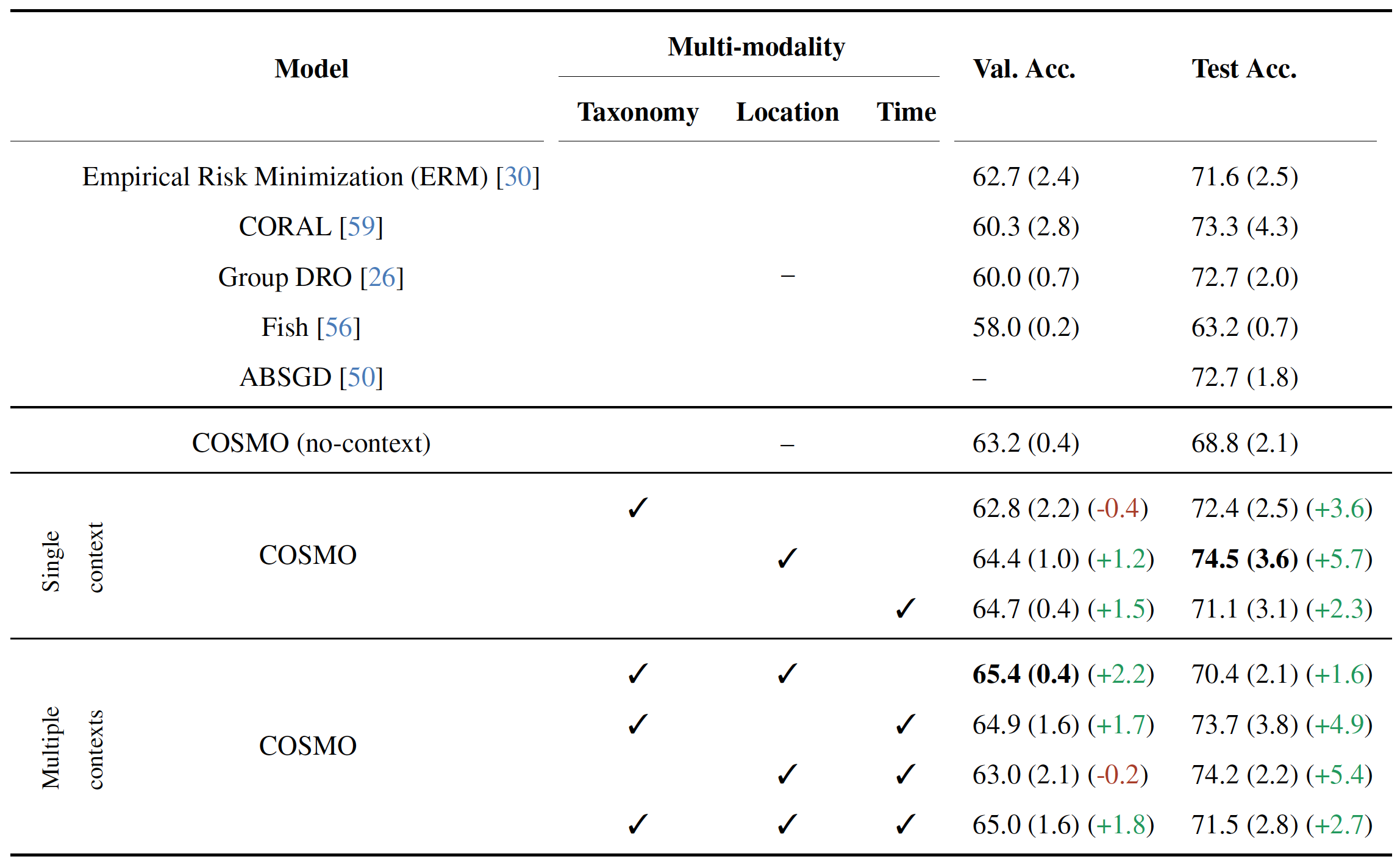
Species Classification results on iWildCam2020-WILDS (OOD) dataset. The first baseline in the second section shows the no-context baseline that uses only image-species labels as KG edges. All models use a pre-trained ResNet-50 as image encoder. Parentheses show standard deviation across 3 random seeds. Missing values are denoted by –.
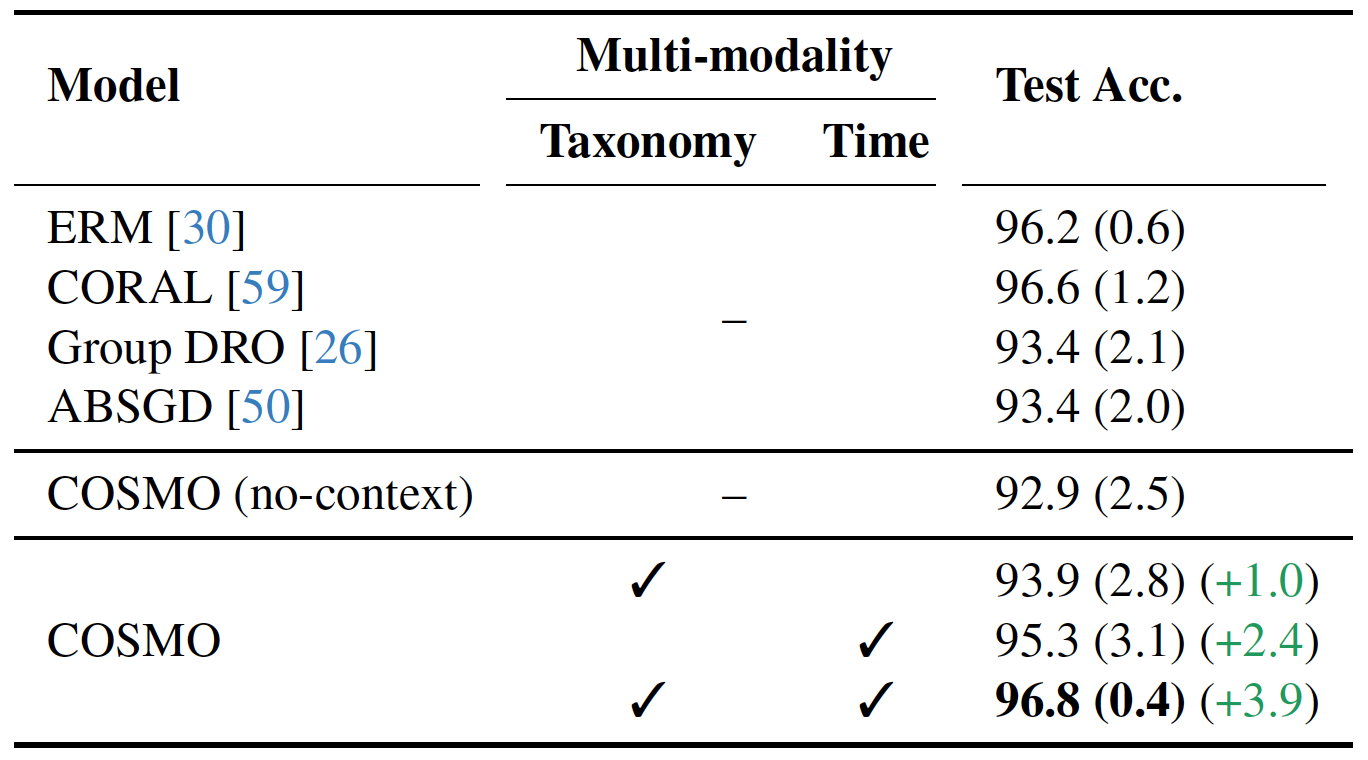
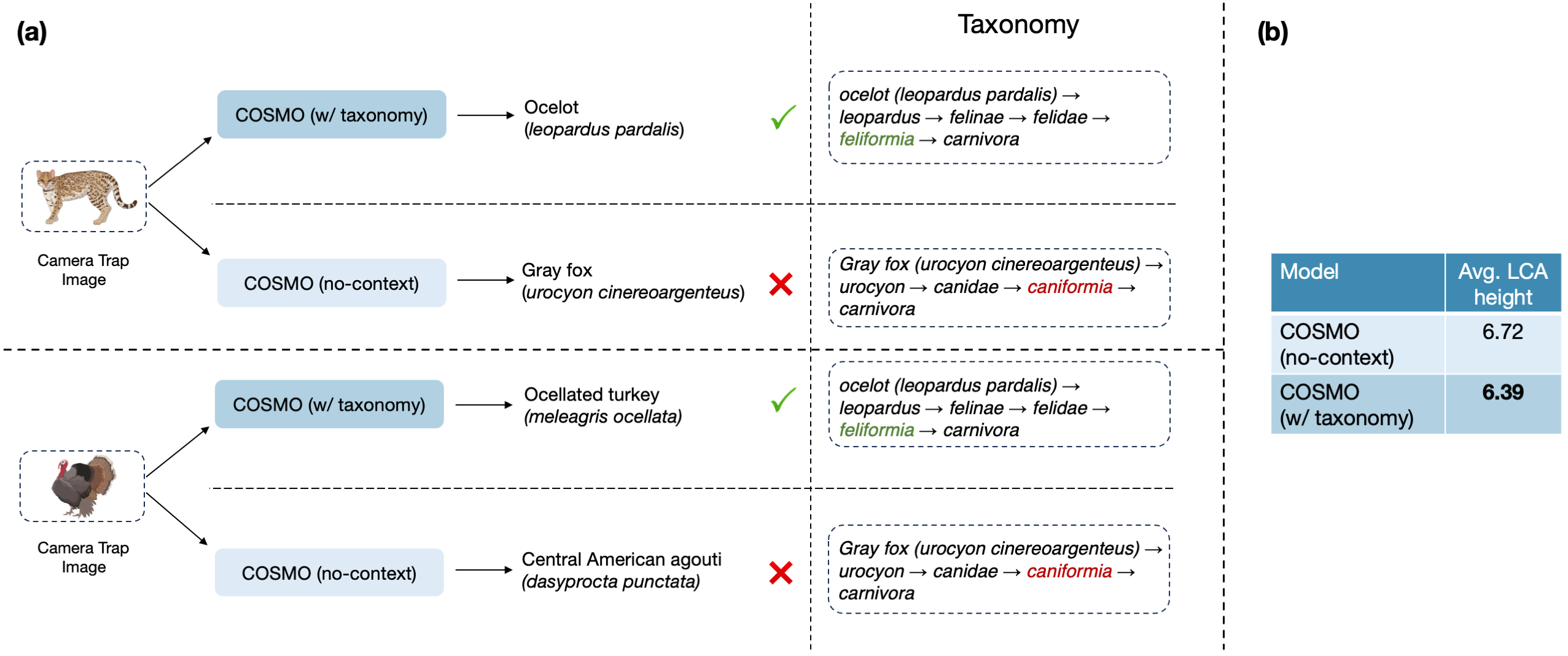
Species Classification results on Snapshot Mountain Zebra dataset.
@article{pahuja2023bringing,
title={Bringing Back the Context: Camera Trap Species Identification as Link Prediction on Multimodal Knowledge Graphs},
author={Pahuja, Vardaan and Luo, Weidi and Gu, Yu and Tu, Cheng-Hao and Chen, Hong-You and Berger-Wolf, Tanya and Stewart, Charles and Gao, Song and Chao, Wei-Lun and Su, Yu},
journal={arXiv preprint arXiv:2401.00608},
year={2023}
}